Mechanism Supports Function: The Dharmacon™ siRNA Algorithm 20 Years Strong
RNA interference using small interfering RNAs (siRNAs) has become a mainstay of functional gene characterization and has generated over a dozen FDA-approved therapeutics and drugs in late-stage clinical trials. Algorithms for selection of functional siRNA sequences were developed early on; however, some determinants for rational selection of siRNAs have remained elusive until recently. A mechanistic study by Peter Wang in David Bartel's lab sheds new light on slicing kinetics of the Argonaute enzyme central to RNA interference.
Early siRNA Design Principles
Soon after siRNAs were discovered in the early 2000s, they were quickly harnessed as research tools enabling the growing field of functional genomics. Rules had to be established for selecting potent siRNAs to turn this discovery into useful technology: small RNA-directed knockdown of nearly any desired transcript. Early studies of siRNAs and microRNAs allowed for both rational design and machine learning-assisted selection of functional molecules.
Many features of potent siRNA duplexes were first published in 2004 by scientists at Dharmacon,Inc. (Reynolds, 2004). Some observations could be described by the known function of Argonaute 2 (Ago2), the main effector of the RNA-Induced Silencing Complex (RISC) in mammalian cells. For instance, both low G/C content and low internal stability at the "sense" strand 3′-terminus aid Ago2 in unwinding the siRNA duplex and selecting the correct targeting strand. However, some functional determinants were less easily explained, such as base preferences at certain positions in the duplex (positions 3, 10, and 13 in the sense strand, corresponding to positions 7, 10, and 17 in the targeting strand).
New Mechanistic Insights
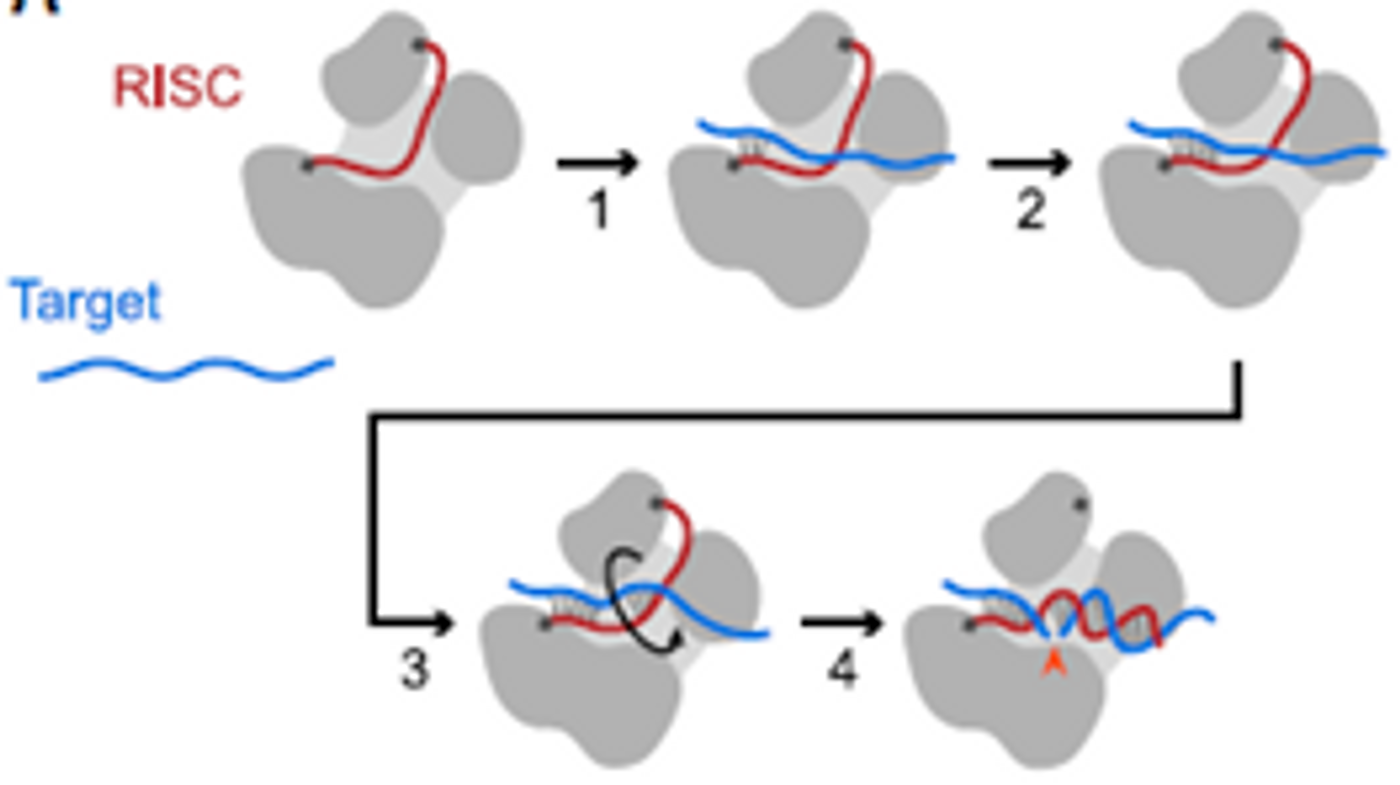
As a result of the work showcased in Wang, 2024, a more complete picture has emerged of the dynamic Ago2 complex. Slicing can now be explained as a 4-step model involving binding in the seed region of the targeting strand, propagation through the seed, a second nucleation event to center the slicing target, and a helical rotation to complete base pairing throughout the site. Slicing kinetics are affected by intermediate steps (including rearrangement inside Ago2) to form a complete duplex between the targeting strand and target mRNA.
Figure 1 adapted from Wang, 2024 [published under CC-BY license]
The machine-learning approach of the original Dharmacon algorithm had predicted cryptic preferences of an A at position 3, a U at position 10, and absence of G at position 13, but could not rationally explain why these were beneficial for function. Thanks to this detailed biochemical study, we now understand these design elements from a biophysical perspective, confirming that Dharmacon siRNA design remains state-of-the-art for potent gene knockdown.
Explore Dharmacon siRNA Solutions
Discover how Revvity's proven Dharmacon siRNA technology can advance your research. Visit https://horizondiscovery.com/en/gene-modulation/knockdown/sirna to explore our complete range of siRNA solutions for functional genomics and therapeutic development.
References
Wang PY, Bartel DP. The guide-RNA sequence dictates the slicing kinetics and conformational dynamics of the Argonaute silencing complex. Mol Cell. 2024 Aug 8;84(15):2918-2934.e11. doi: 10.1016/j.molcel.2024.06.026.
Reynolds, A. et al. Rational design for RNA interference. Nature Biotechnol. 2004 Feb 1 doi:10.1038/nbt936.